卷积神经网络(CNN):乳腺癌识别.ipynb
文章目录
- 一、前言
- 一、设置GPU
- 二、导入数据
- 1. 导入数据
- 2. 检查数据
- 3. 配置数据集
- 4. 数据可视化
- 三、构建模型
- 四、编译
- 五、训练模型
- 六、评估模型
- 1. Accuracy与Loss图
- 2. 混淆矩阵
- 3. 各项指标评估
一、前言
我的环境:
- 语言环境:Python3.6.5
- 编译器:jupyter notebook
- 深度学习环境:TensorFlow2.4.1
往期精彩内容:
- 卷积神经网络(CNN)实现mnist手写数字识别
- 卷积神经网络(CNN)多种图片分类的实现
- 卷积神经网络(CNN)衣服图像分类的实现
- 卷积神经网络(CNN)鲜花识别
- 卷积神经网络(CNN)天气识别
- 卷积神经网络(VGG-16)识别海贼王草帽一伙
- 卷积神经网络(ResNet-50)鸟类识别
- 卷积神经网络(AlexNet)鸟类识别
- 卷积神经网络(CNN)识别验证码
来自专栏:机器学习与深度学习算法推荐
一、设置GPU
import tensorflow as tf
gpus = tf.config.list_physical_devices("GPU")
if gpus:
gpu0 = gpus[0] #如果有多个GPU,仅使用第0个GPU
tf.config.experimental.set_memory_growth(gpu0, True) #设置GPU显存用量按需使用
tf.config.set_visible_devices([gpu0],"GPU")
import matplotlib.pyplot as plt
import os,PIL,pathlib
import numpy as np
import pandas as pd
import warnings
from tensorflow import keras
warnings.filterwarnings("ignore") #忽略警告信息
plt.rcParams['font.sans-serif'] = ['SimHei'] # 用来正常显示中文标签
plt.rcParams['axes.unicode_minus'] = False # 用来正常显示负号
二、导入数据
1. 导入数据
import pathlib
data_dir = "./32-data"
data_dir = pathlib.Path(data_dir)
image_count = len(list(data_dir.glob('*/*')))
print("图片总数为:",image_count)
图片总数为: 13403
batch_size = 16
img_height = 50
img_width = 50
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.2,
subset="training",
seed=12,
image_size=(img_height, img_width),
batch_size=batch_size)
Found 13403 files belonging to 2 classes.
Using 10723 files for training.
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.2,
subset="validation",
seed=12,
image_size=(img_height, img_width),
batch_size=batch_size)
Found 13403 files belonging to 2 classes.
Using 2680 files for validation.
class_names = train_ds.class_names
print(class_names)
['0', '1']
2. 检查数据
for image_batch, labels_batch in train_ds:
print(image_batch.shape)
print(labels_batch.shape)
break
(16, 50, 50, 3)
(16,)
3. 配置数据集
AUTOTUNE = tf.data.AUTOTUNE
def train_preprocessing(image,label):
return (image/255.0,label)
train_ds = (
train_ds.cache()
.shuffle(1000)
.map(train_preprocessing) # 这里可以设置预处理函数
# .batch(batch_size) # 在image_dataset_from_directory处已经设置了batch_size
.prefetch(buffer_size=AUTOTUNE)
)
val_ds = (
val_ds.cache()
.shuffle(1000)
.map(train_preprocessing) # 这里可以设置预处理函数
# .batch(batch_size) # 在image_dataset_from_directory处已经设置了batch_size
.prefetch(buffer_size=AUTOTUNE)
)
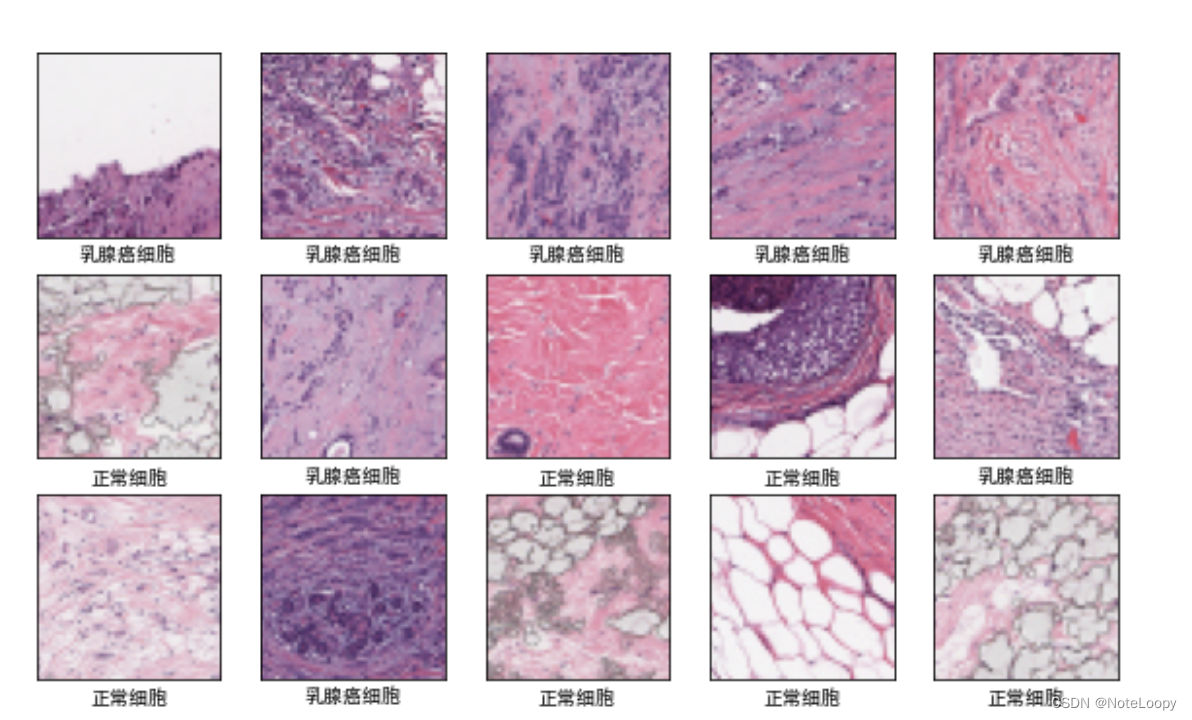
4. 数据可视化
plt.figure(figsize=(10, 8)) # 图形的宽为10高为5
plt.suptitle("数据展示")
class_names = ["乳腺癌细胞","正常细胞"]
for images, labels in train_ds.take(1):
for i in range(15):
plt.subplot(4, 5, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
# 显示图片
plt.imshow(images[i])
# 显示标签
plt.xlabel(class_names[labels[i]-1])
plt.show()
三、构建模型
import tensorflow as tf
model = tf.keras.Sequential([
tf.keras.layers.Conv2D(filters=16,kernel_size=(3,3),padding="same",activation="relu",input_shape=[img_width, img_height, 3]),
tf.keras.layers.Conv2D(filters=16,kernel_size=(3,3),padding="same",activation="relu"),
tf.keras.layers.MaxPooling2D((2,2)),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Conv2D(filters=16,kernel_size=(3,3),padding="same",activation="relu"),
tf.keras.layers.MaxPooling2D((2,2)),
tf.keras.layers.Conv2D(filters=16,kernel_size=(3,3),padding="same",activation="relu"),
tf.keras.layers.MaxPooling2D((2,2)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(2, activation="softmax")
])
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv2d (Conv2D) (None, 50, 50, 16) 448
_________________________________________________________________
conv2d_1 (Conv2D) (None, 50, 50, 16) 2320
_________________________________________________________________
max_pooling2d (MaxPooling2D) (None, 25, 25, 16) 0
_________________________________________________________________
dropout (Dropout) (None, 25, 25, 16) 0
_________________________________________________________________
conv2d_2 (Conv2D) (None, 25, 25, 16) 2320
_________________________________________________________________
max_pooling2d_1 (MaxPooling2 (None, 12, 12, 16) 0
_________________________________________________________________
conv2d_3 (Conv2D) (None, 12, 12, 16) 2320
_________________________________________________________________
max_pooling2d_2 (MaxPooling2 (None, 6, 6, 16) 0
_________________________________________________________________
flatten (Flatten) (None, 576) 0
_________________________________________________________________
dense (Dense) (None, 2) 1154
=================================================================
Total params: 8,562
Trainable params: 8,562
Non-trainable params: 0
_________________________________________________________________
四、编译
model.compile(optimizer="adam",
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
五、训练模型
from tensorflow.keras.callbacks import ModelCheckpoint, Callback, EarlyStopping, ReduceLROnPlateau, LearningRateScheduler
NO_EPOCHS = 100
PATIENCE = 5
VERBOSE = 1
# 设置动态学习率
annealer = LearningRateScheduler(lambda x: 1e-3 * 0.99 ** (x+NO_EPOCHS))
# 设置早停
earlystopper = EarlyStopping(monitor='loss', patience=PATIENCE, verbose=VERBOSE)
#
checkpointer = ModelCheckpoint('best_model.h5',
monitor='val_accuracy',
verbose=VERBOSE,
save_best_only=True,
save_weights_only=True)
train_model = model.fit(train_ds,
epochs=NO_EPOCHS,
verbose=1,
validation_data=val_ds,
callbacks=[earlystopper, checkpointer, annealer])
六、评估模型
1. Accuracy与Loss图
acc = train_model.history['accuracy']
val_acc = train_model.history['val_accuracy']
loss = train_model.history['loss']
val_loss = train_model.history['val_loss']
epochs_range = range(len(acc))
plt.figure(figsize=(12, 4))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label='Training Accuracy')
plt.plot(epochs_range, val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.title('Training and Validation Accuracy')
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label='Training Loss')
plt.plot(epochs_range, val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.title('Training and Validation Loss')
plt.show()
2. 混淆矩阵
from sklearn.metrics import confusion_matrix
import seaborn as sns
import pandas as pd
# 定义一个绘制混淆矩阵图的函数
def plot_cm(labels, predictions):
# 生成混淆矩阵
conf_numpy = confusion_matrix(labels, predictions)
# 将矩阵转化为 DataFrame
conf_df = pd.DataFrame(conf_numpy, index=class_names ,columns=class_names)
plt.figure(figsize=(8,7))
sns.heatmap(conf_df, annot=True, fmt="d", cmap="BuPu")
plt.title('混淆矩阵',fontsize=15)
plt.ylabel('真实值',fontsize=14)
plt.xlabel('预测值',fontsize=14)
val_pre = []
val_label = []
for images, labels in val_ds:#这里可以取部分验证数据(.take(1))生成混淆矩阵
for image, label in zip(images, labels):
# 需要给图片增加一个维度
img_array = tf.expand_dims(image, 0)
# 使用模型预测图片中的人物
prediction = model.predict(img_array)
val_pre.append(class_names[np.argmax(prediction)])
val_label.append(class_names[label])
plot_cm(val_label, val_pre)
3. 各项指标评估
from sklearn import metrics
def test_accuracy_report(model):
print(metrics.classification_report(val_label, val_pre, target_names=class_names))
score = model.evaluate(val_ds, verbose=0)
print('Loss function: %s, accuracy:' % score[0], score[1])
test_accuracy_report(model)
precision recall f1-score support
乳腺癌细胞 0.92 0.90 0.91 1339
正常细胞 0.91 0.92 0.91 1341
accuracy 0.91 2680
macro avg 0.91 0.91 0.91 2680
weighted avg 0.91 0.91 0.91 2680
Loss function: 0.22688131034374237, accuracy: 0.9138059616088867
pport
乳腺癌细胞 0.92 0.90 0.91 1339
正常细胞 0.91 0.92 0.91 1341
accuracy 0.91 2680
macro avg 0.91 0.91 0.91 2680
weighted avg 0.91 0.91 0.91 2680
Loss function: 0.22688131034374237, accuracy: 0.9138059616088867
